Register for free to join our community of investors and share your ideas. You will also get access to streaming quotes, interactive charts, trades, portfolio, live options flow and more tools.
Lesser IPO restrictions are good for dnag.
http://www.lexology.com/library/detail.aspx?g=c2bcddb8-2e59-4c61-9e25-d8b607fa6a03
ok looks like 0007 close. have a nice weekend.
any big news and easy 10 bagger from here.
agree, 250%+ is just great.
7 close 2day. The run is still going strong. 00s next week.
time for eod volume. take out 0007 13min left
could be. hope so.
I bet good news is coming soon with this much activity. I'm buying more while I can and hanging for the long ride back up!
last DNGA news from 2009. 3 years ago. time for something big.
DNAG is a shell stock. R/M news would be great.
cant find any news. perhaps monday. 41 milion trading volume lookgin great
time to break 0007 with major hits
13.2 milion bid support
UBSS NITE ETMM CSTI left @0007. players hold hour shares. only a fake fall
power hour start now. 0007 break coming. just 1.3 milion left and we create a new hood
15 milion bid HUGÈEEEEEEEEEEEE
12 milion bid 1 milion ask
0007 up big printing
0006 up big hitting
going back up
Rumors are that one group will be coming in around 3:30
0004x0005 500k left and 0007 back up
1x0005 left and 0004 bid 11 milion wow
1x 0005 cheap shares then 0007 up
after 0007 we can go big
great trading volume today. great
Just gets better in here, huge bid. Ready for a move eod imo
agree, big move coming before close. and monday just up up up.. DNAG monday on radar.
monday big gapper for sure.
28.5 milion volume and great accumulation 0007. great sound and ready to go into.
9x more bid than ask just wowwwwwwwwwwwwwww
this thing is out off control.. will blow into
0007 on fire 0008 and big pop coming . nothing left
|
Followers
|
297
|
Posters
|
|
|
Posts (Today)
|
0
|
Posts (Total)
|
82595
|
|
Created
|
08/19/00
|
Type
|
Free
|
| Moderators | |||
DNA
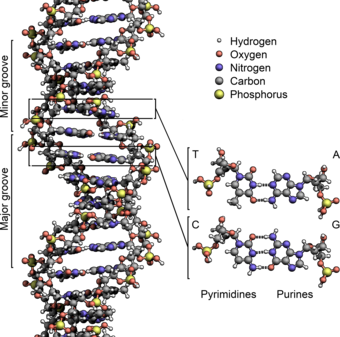

Deoxyribonucleic acid (DNA) is a molecule encoding the genetic instructions used in the development and functioning of all known living organisms and many viruses. Along with RNA and proteins, DNA is one of the three major macromolecules that are essential for all known forms of life. Genetic information is encoded as a sequence of nucleotides (guanine, adenine, thymine, and cytosine) recorded using the letters G, A, T, and C. Most DNA molecules are double-stranded helices, consisting of two long polymers of simple units called nucleotides, molecules with backbones made of alternating sugars (deoxyribose) and phosphate groups (related to phosphoric acid), with the nucleobases (G, A, T, C) attached to the sugars. DNA is well-suited for biological information storage, since the DNA backbone is resistant to cleavage and the double-stranded structure provides the molecule with a built-in duplicate of the encoded information.
These two strands run in opposite directions to each other and are therefore anti-parallel, one backbone being 3' (three prime) and the other 5' (five prime). This refers to the direction the 3rd and 5th carbon on the sugar molecule is facing. Attached to each sugar is one of four types of molecules called nucleobases (informally, bases). It is the sequence of these four nucleobases along the backbone that encodes information. This information is read using the genetic code, which specifies the sequence of the amino acids within proteins. The code is read by copying stretches of DNA into the related nucleic acid RNA in a process called transcription.
Within cells, DNA is organized into long structures called chromosomes. During cell division these chromosomes are duplicated in the process of DNA replication, providing each cell its own complete set of chromosomes. Eukaryotic organisms (animals, plants, fungi, and protists) store most of their DNA inside the cell nucleus and some of their DNA in organelles, such as mitochondria or chloroplasts.[1] In contrast, prokaryotes (bacteria and archaea) store their DNA only in the cytoplasm. Within the chromosomes, chromatin proteins such as histones compact and organize DNA. These compact structures guide the interactions between DNA and other proteins, helping control which parts of the DNA are transcribed.
Properties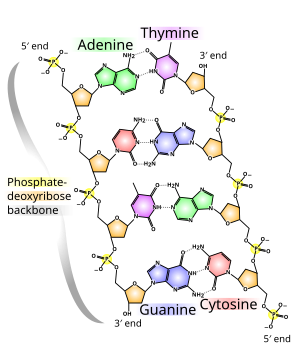
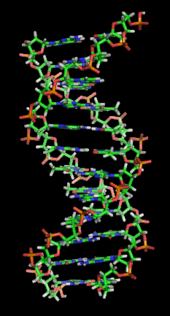
DNA is a long polymer made from repeating units called nucleotides.[2][3][4] DNA was first identified and isolated by Friedrich Miescher and the double helix structure of DNA was first discovered by James D. Watson and Francis Crick. The structure of DNA of all species comprises two helical chains each coiled round the same axis, and each with a pitch of 34 ångströms (3.4 nanometres) and a radius of 10 ångströms (1.0 nanometres).[5] According to another study, when measured in a particular solution, the DNA chain measured 22 to 26 ångströms wide (2.2 to 2.6 nanometres), and one nucleotide unit measured 3.3 Å (0.33 nm) long.[6] Although each individual repeating unit is very small, DNA polymers can be very large molecules containing millions of nucleotides. For instance, the largest human chromosome, chromosome number 1, is approximately 220 million base pairs long.[7]
In living organisms DNA does not usually exist as a single molecule, but instead as a pair of molecules that are held tightly together.[8][9] These two long strands entwine like vines, in the shape of a double helix. The nucleotide repeats contain both the segment of the backbone of the molecule, which holds the chain together, and a nucleobase, which interacts with the other DNA strand in the helix. A nucleobase linked to a sugar is called a nucleoside and a base linked to a sugar and one or more phosphate groups is called a nucleotide. A polymer comprising multiple linked nucleotides (as in DNA) is called a polynucleotide.[10]
The backbone of the DNA strand is made from alternating phosphate and sugar residues.[11] The sugar in DNA is 2-deoxyribose, which is a pentose (five-carbon) sugar. The sugars are joined together by phosphate groups that form phosphodiester bonds between the third and fifth carbon atoms of adjacent sugar rings. These asymmetric bonds mean a strand of DNA has a direction. In a double helix the direction of the nucleotides in one strand is opposite to their direction in the other strand: the strands are antiparallel. The asymmetric ends of DNA strands are called the 5′ (five prime) and 3′ (three prime) ends, with the 5' end having a terminal phosphate group and the 3' end a terminal hydroxyl group. One major difference between DNA and RNA is the sugar, with the 2-deoxyribose in DNA being replaced by the alternative pentose sugar ribose in RNA.[9]
The DNA double helix is stabilized primarily by two forces: hydrogen bonds between nucleotides and base-stacking interactions among aromatic nucleobases.[13] In the aqueous environment of the cell, the conjugated π bonds of nucleotide bases align perpendicular to the axis of the DNA molecule, minimizing their interaction with the solvation shell and therefore, the Gibbs free energy. The four bases found in DNA are adenine (abbreviated A), cytosine (C), guanine (G) and thymine (T). These four bases are attached to the sugar/phosphate to form the complete nucleotide, as shown for adenosine monophosphate.
The nucleobases are classified into two types: the purines, A and G, being fused five- and six-membered heterocyclic compounds, and the pyrimidines, the six-membered rings C and T.[9] A fifth pyrimidine nucleobase, uracil (U), usually takes the place of thymine in RNA and differs from thymine by lacking a methyl group on its ring. In addition to RNA and DNA a large number of artificial nucleic acid analogues have also been created to study the properties of nucleic acids, or for use in biotechnology.[14]
Uracil is not usually found in DNA, occurring only as a breakdown product of cytosine. However in a number of bacteriophages - Bacillus subtilis bacteriophages PBS1 and PBS2 and Yersinia bacteriophage piR1-37 - thymine has been replaced by uracil.[15] A modified form (beta-d-glucopyranosyloxymethyluracil) is also found in a number of organisms: the flagellates Diplonema and Euglena, and all the kinetoplastid genera[16] Biosynthesis of J occurs in two steps: in the first step a specific thymidine in DNA is converted into hydroxymethyldeoxyuridine; in the second HOMedU is glycosylated to form J.[17] Proteins that bind specifically to this base have been identified.[18][19][20] These proteins appear to be distant relatives of the Tet1 oncogene that is involved in the pathogenesis of acute myeloid leukemia.[21] J appears to act as a termination signal for RNA polymerase II.[22][23]
| Volume | |
| Day Range: | |
| Bid Price | |
| Ask Price | |
| Last Trade Time: |
